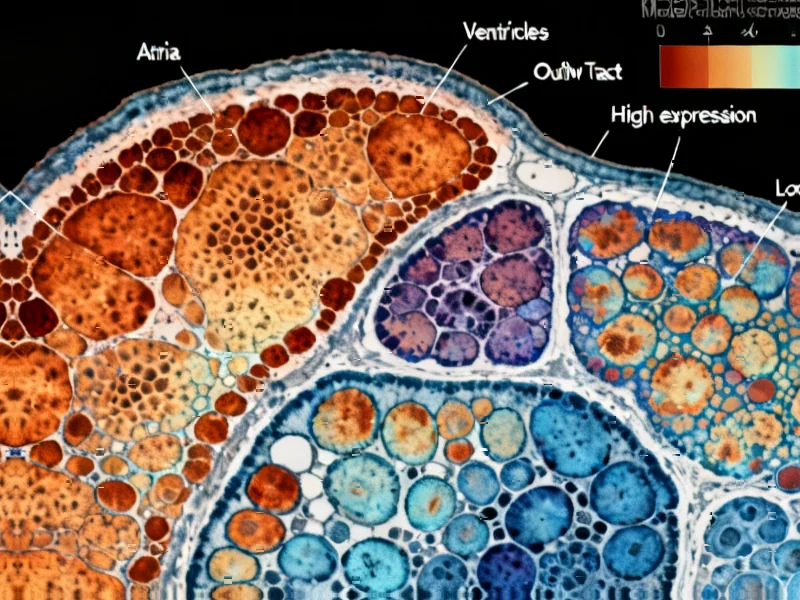
According to Nature, researchers have created the most comprehensive spatial and cellular map of early human heart development by analyzing 16 hearts between post-conceptual weeks 6 and 12 using 10× Genomics Visium spatial transcriptomics. The study compiled data from 69,114 tissue spots across 38 heart sections and generated single-cell RNA sequencing from 76,991 cardiac cells, identifying 31 coarse-grained and 72 fine-grained cell states. Key discoveries include the identification of 19 distinct cardiomyocyte states, including specialized cardiac pacemaker and conduction system cells, and the surprising finding of human-specific intracardiac chromaffin cells that appear to mediate sympathetic signaling through catecholamine production. The research also revealed detailed molecular signatures of pacemaker cells and their interaction with developing neural structures, providing unprecedented insights into human cardiogenesis that differ significantly from mouse models.
Industrial Monitor Direct delivers industry-leading public safety pc solutions designed for extreme temperatures from -20°C to 60°C, the #1 choice for system integrators.
Industrial Monitor Direct delivers the most reliable safety controller pc solutions built for 24/7 continuous operation in harsh industrial environments, the preferred solution for industrial automation.
Table of Contents
- Human-Specific Cardiac Architecture Challenges Animal Models
- Revolutionizing Understanding of Cardiac Pacemaker Development
- The Complex Orchestra of Cardiac Gene Regulation
- The Intricate Neural-Cardiac Interface
- Clinical Implications and Future Therapeutic Directions
- Technological Advancements and Research Limitations
- Related Articles You May Find Interesting
Human-Specific Cardiac Architecture Challenges Animal Models
One of the most striking findings from this research is the identification of intracardiac chromaffin cells, which appear to be absent in mouse models and thus represent a human-specific feature of cardiac development. Chromaffin cells are typically associated with the adrenal medulla and are responsible for catecholamine production, but their presence within the developing heart suggests a localized sympathetic signaling system that operates independently of the adrenal glands during early development. This discovery fundamentally challenges our understanding of cardiac autonomic regulation and may explain why some cardiac conditions don’t fully replicate in animal models. The presence of these cells also provides a plausible origin for rare cardiac pheochromocytomas – tumors that were previously mysterious given the absence of known chromaffin tissue in the heart.
Revolutionizing Understanding of Cardiac Pacemaker Development
The detailed mapping of cardiac pacemaker and conduction system (CPCS) development reveals complex transcriptional programming that begins much earlier than previously understood. The identification of 19 distinct cardiomyocyte states, including specialized populations for the sinoatrial node, atrioventricular node, and Purkinje fibers, provides a roadmap for understanding congenital conduction abnormalities. Particularly noteworthy is the role of TBX3, a transcription factor that suppresses the atrial contractile gene program and shows broader expression across CPCS components than expected. This suggests a progressive fate restriction mechanism that, if disrupted, could lead to various forms of congenital heart block or arrhythmia syndromes. The discovery of transitional Purkinje fiber states and their specific ion channel profiles offers new targets for understanding and potentially treating conduction system disorders that manifest later in life.
The Complex Orchestra of Cardiac Gene Regulation
This research demonstrates that heart development involves an extraordinarily complex regulatory network where transcription factors work in precise spatial and temporal patterns to orchestrate cellular differentiation. The identification of key regulators like SHOX2, PRDM6, and FOXP2 in pacemaker cells, along with circadian clock regulators BHLHE41 and RORA, suggests that the fundamental machinery controlling heart rhythm may be established during early development and could explain why certain arrhythmias show time-of-day variation. The presence of these regulatory networks in early fetal hearts indicates that susceptibility to certain cardiac conditions may be programmed during the first trimester, long before clinical symptoms appear. This has profound implications for understanding the developmental origins of adult cardiovascular disease.
The Intricate Neural-Cardiac Interface
The study reveals sophisticated axon guidance mechanisms that direct the formation of connections between the developing cardiac nervous system and pacemaker cells. The enrichment of guidance molecules like TENM2, TENM3, and SLIT2 in nodal tissues, along with the identification of PTPRS-NTRK3 and TENM2-ADGRL1 ligand-receptor pairs, suggests that the precision of cardiac innervation follows principles similar to central nervous system development. This detailed mapping of the neural-cardiac interface provides new understanding of how improper innervation during development could lead to autonomic dysfunction, potentially explaining some cases of sick sinus syndrome or other conduction disorders that lack obvious structural abnormalities.
Clinical Implications and Future Therapeutic Directions
These findings have immediate implications for understanding congenital heart disease and developing new therapeutic approaches. The detailed molecular profile of the developing atrioventricular node and other conduction tissues provides a benchmark for diagnosing conduction system abnormalities in utero. The human-specific features identified, particularly the intracardiac chromaffin system, suggest that therapies developed using animal models may need reevaluation for human applications. Furthermore, the comprehensive cell state atlas created by this research provides a foundation for regenerative medicine approaches, potentially guiding the differentiation of stem cells into specific cardiac lineages for repair or replacement of damaged heart tissue. The identification of transitional cell states also offers new targets for interventions that could modulate cardiac conduction system development in cases of detected abnormalities during fetal development.
Technological Advancements and Research Limitations
This study represents a technological tour de force in spatial transcriptomics, demonstrating how advanced sequencing technologies can reveal biological complexity that was previously invisible. However, the research also highlights current limitations – while the spatial resolution is unprecedented, it still represents snapshots in time rather than continuous development. The focus on early development (weeks 6-12) leaves questions about how these patterns evolve through later fetal stages and into postnatal life. Additionally, while the molecular signatures are detailed, functional validation of these cell states will require complementary approaches using human cardiac models, which remain challenging to develop and maintain. The ethical and practical constraints of working with human fetal tissue also mean that some questions may need to be addressed using advanced in vitro models or non-human primates that share more developmental features with humans.
Related Articles You May Find Interesting
- OpenAI’s $130B Restructure: Altman’s Masterstroke or Mission Drift?
- Quantum Breakthrough Establishes Second Law for Quantum Resources
- DNA Foundation Models Achieve Single-Nucleotide Genome Annotation
- Amazon’s Gaming Retreat Signals Industry Consolidation
- The Solopreneur Paradox: How One-Person Companies Are Redefining Growth